-Search query
-Search result
Showing 1 - 50 of 81 items for (author: kossiakoff & aa)

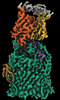
EMDB-19440: 
Cryo-EM structure of human NTCP-Bulevirtide complex
Method: single particle / : Liu H, Zakrzewicz D, Nosol K, Irobalieva RN, Mukherjee S, Bang-Soerensen R, Goldmann N, Kunz S, Rossi L, Kossiakoff AA, Urban S, Glebe D, Geyer J, Locher KP
EMDB-41255: 
Cryo-EM structure of Pseudomonas aeruginosa TonB-dependent transporter PhuR in complex with synthetic antibody and heme
Method: single particle / : Knejski P, Erramilli SK, Kossiakoff AA

EMDB-17655: 
Human OATP1B3
Method: single particle / : Ciuta AD, Nosol K, Kowal J, Mukherjee S, Ramirez AS, Stieger B, Kossiakoff AA, Locher KP

EMDB-17677: 
Human OATP1B1
Method: single particle / : Ciuta AD, Nosol K, Kowal J, Mukherjee S, Ramirez AS, Stieger B, Kossiakoff AA, Locher KP

EMDB-27674: 
LRRC8A:C conformation 2 (oblong) top mask
Method: single particle / : Kern DM, Brohawn SG

EMDB-27675: 
LRRC8A:C conformation 2 (oblong) LRR mask
Method: single particle / : Kern DM, Brohawn SG

EMDB-27676: 
LRRC8A:C conformation 2 (oblong)
Method: single particle / : Kern DM, Brohawn SG

EMDB-27677: 
LRRC8A:C conformation 1 (round) top focus
Method: single particle / : Kern DM, Brohawn SG

EMDB-27678: 
LRRC8A:C conformation 1 (round) LRR focus 1
Method: single particle / : Kern DM, Brohawn SG

EMDB-27679: 
LRRC8A:C conformation 1 (round) LRR focus 2
Method: single particle / : Kern DM, Brohawn SG

EMDB-27681: 
LRRC8A:C conformation 1 (round) LRR focus 3
Method: single particle / : Kern DM, Brohawn SG

EMDB-27682: 
LRRC8A:C conformation 1 (round)
Method: single particle / : Kern DM, Brohawn SG

EMDB-27686: 
LRRC8A:C in MSPE3D1 nanodisc top focus
Method: single particle / : Kern DM, Brohawn SG

EMDB-27687: 
LRRC8A:C in MSP1E3D1 nanodisc
Method: single particle / : Kern DM, Brohawn SG

EMDB-28894: 
LRRC8A(T48D):C conformation 2 top focus
Method: single particle / : Kern DM, Brohawn SG

EMDB-28895: 
LRRC8A(T48D):C conformation 2 LRR focus
Method: single particle / : Kern DM, Brohawn SG

EMDB-28897: 
LRRC8A(T48D):C conformation 2 top focus
Method: single particle / : Kern DM, Brohawn SG

EMDB-28898: 
LRRC8A(T48D):C conformation 2 top focus
Method: single particle / : Kern DM, Brohawn SG

EMDB-28901: 
LRRC8A(T48D):C conformation 2 top focus
Method: single particle / : Kern DM, Brohawn SG

EMDB-28903: 
LRRC8A(T48D):C conformation 2 top focus
Method: single particle / : Kern DM, Brohawn SG

EMDB-28904: 
LRRC8A(T48D):C conformation 2 top focus
Method: single particle / : Kern DM, Brohawn SG

EMDB-28905: 
LRRC8A(T48D):C conformation 2 top focus
Method: single particle / : Kern DM, Brohawn SG

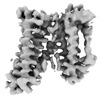
EMDB-14779: 
Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with acceptor peptide and bound to CMT2-Fab and anti-Fab nanobody
Method: single particle / : Bloch JS, Mukherjee S, Mao R, Irobalieva R, Kossiakoff AA, Goddard-Borger ED, Locher KP

EMDB-14780: 
Cryo-EM structure of C-mannosyltransferase CeDPY19, in apo state, bound to CMT2-Fab and anti-Fab nanobody
Method: single particle / : Bloch JS, Mukherjee S, Irobalieva R, Kossiakoff AA, Goddard-Borger ED, Locher KP

EMDB-14781: 
Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with Dol25-P-Man and bound to CMT2-Fab and anti-Fab nanobody
Method: single particle / : Bloch JS, Mukherjee S, Boilevin J, Irobalieva R, Darbre T, Reymond JL, Kossiakoff AA, Goddard-Borger ED, Locher KP

EMDB-14782: 
Cryo-EM structure of C-mannosyltransferase CeDPY19, in ternary complex with Dol25-P-C-Man and acceptor peptide, bound to CMT2-Fab and anti-Fab nanobody
Method: single particle / : Bloch JS, Mao R, Mukherjee S, Boilevin J, Irobalieva R, Darbre T, Reymond JL, Kossiakoff AA, Goddard-Borger ED, Locher KP
EMDB-24698: 
Cryo-EM structure of the HIV-1 restriction factor human SERINC3
Method: single particle / : Purdy MD, Leonhardt SA, Yeager M

EMDB-24705: 
Human SERINC3-DeltaICL4
Method: single particle / : Purdy MD, Leonhardt SA, Yeager M

EMDB-25836: 
CryoEM Structure of sFab COP-3 Complex with human claudin-4 and Clostridium perfringens enterotoxin C-terminal domain focused map
Method: single particle / : Vecchio AJ, Orlando BJ
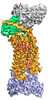
EMDB-25171: 
Cryo-EM structure of ACKR3 in complex with CXCL12, an intracellular Fab, and an extracellular Fab
Method: single particle / : Yen YC, Schafer CT, Gustavsson M, Handel TM, Tesmer JJG

EMDB-25172: 
Cryo-EM structure of ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ, an intracellular Fab, and an extracellular Fab
Method: single particle / : Yen YC, Schafer CT, Gustavsson M, Handel TM, Tesmer JJG
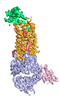
EMDB-25173: 
Cryo-EM structure of ACKR3 in complex with CXCL12 and an intracellular Fab
Method: single particle / : Yen YC, Schafer CT, Gustavsson M, Handel TM, Tesmer JJG
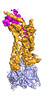
EMDB-25174: 
Cryo-EM structure of human ACKR3 in complex with chemokine N-terminal mutant CXCL12_LRHQ and an intracellular Fab
Method: single particle / : Yen YC, Schafer CT, Gustavsson M, Handel TM, Tesmer JJG
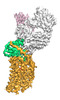
EMDB-25175: 
Cryo-EM structure of human ACKR3 in complex with CXCL12, a small molecule partial agonist CCX662, and an extracellular Fab
Method: single particle / : Yen YC, Schafer CT, Gustavsson M, Handel TM, Tesmer JJG
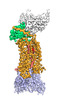
EMDB-25176: 
Cryo-EM structure of human ACKR3 in complex with CXCL12, a small molecule partial agonist CCX662, an extracellular Fab, and an intracellular Fab
Method: single particle / : Yen YC, Schafer CT, Gustavsson M, Handel TM, Tesmer JJG
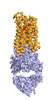
EMDB-25177: 
Cryo-EM structure of human ACKR3 in complex with a small molecule partial agonist CCX662, and an intracellular Fab
Method: single particle / : Yen YC, Schafer CT, Gustavsson M, Handel TM, Tesmer JJG

EMDB-15024: 
Structure of the human sodium/bile acid cotransporter (NTCP) in complex with Fab and nanobody
Method: single particle / : Liu H, Irobalieva RN, Bang-Sorensen R, Nosol K, Mukherjee S, Agrawal P, Stieger B, Kossiakoff AA, Locher KP

EMDB-25750: 
Structure of the peroxisomal retro-translocon formed by a heterotrimeric ubiquitin ligase complex
Method: single particle / : Peiqiang F, Tom R

PDB-7tbl: 
Composite structure of the human nuclear pore complex (NPC) cytoplasmic face generated with a 12A cryo-ET map of the purified HeLa cell NPC
Method: subtomogram averaging / : Bley CJ, Nie S, Mobbs GW, Petrovic S, Gres AT, Liu X, Mukherjee S, Harvey S, Huber FM, Lin DH, Brown B, Tang AW, Rundlet EJ, Correia AR, Chen S, Regmi SG, Stevens TA, Jette CA, Dasso M, Patke A, Palazzo AF, Kossiakoff AA, Hoelz A

PDB-7tbm: 
Composite structure of the dilated human nuclear pore complex (NPC) generated with a 37A in situ cryo-ET map of CD4+ T cell NPC
Method: subtomogram averaging / : Bley CJ, Nie S, Mobbs GW, Petrovic S, Gres AT, Liu X, Mukherjee S, Harvey S, Huber FM, Lin DH, Brown B, Tang AW, Rundlet EJ, Correia AR, Chen S, Regmi SG, Stevens TA, Jette CA, Dasso M, Patke A, Palazzo AF, Kossiakoff AA, Hoelz A
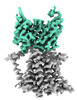
EMDB-26054: 
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state
Method: single particle / : Ashraf KU, Nygaard R, Vickery ON, Erramilli SK, Herrera CM, McConville TH, Petrou VI, Giacometti SI, Dufrisne MB, Nosol K, Zinkle AP, Graham CLB, Loukeris M, Kloss B, Skorupinska-Tudek K, Swiezewska E, Roper D, Clarke OB, Uhlemann AC, Kossiakoff AA, Trent MS, Stansfeld PJ, Mancia F
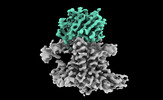
EMDB-26057: 
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state
Method: single particle / : Ashraf KU, Nygaard R, Vickery ON, Erramilli SK, Herrera CM, McConville TH, Petrou VI, Giacometti SI, Dufrisne MB, Nosol K, Zinkle AP, Graham CLB, Loukeris M, Kloss B, Skorupinska-Tudek K, Swiezewska E, Roper D, Clarke OB, Uhlemann AC, Kossiakoff AA, Trent MS, Stansfeld PJ, Mancia F

PDB-7tpg: 
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its ligand bound state
Method: single particle / : Ashraf KU, Nygaard R, Vickery ON, Erramilli SK, Herrera CM, McConville TH, Petrou VI, Giacometti SI, Dufrisne MB, Nosol K, Zinkle AP, Graham CLB, Loukeris M, Kloss B, Skorupinska-Tudek K, Swiezewska E, Roper D, Clarke OB, Uhlemann AC, Kossiakoff AA, Trent MS, Stansfeld PJ, Mancia F

PDB-7tpj: 
Single-Particle Cryo-EM Structure of the WaaL O-antigen ligase in its apo state
Method: single particle / : Ashraf KU, Nygaard R, Vickery ON, Erramilli SK, Herrera CM, McConville TH, Petrou VI, Giacometti SI, Dufrisne MB, Nosol K, Zinkle AP, Graham CLB, Loukeris M, Kloss B, Skorupinska-Tudek K, Swiezewska E, Roper D, Clarke OB, Uhlemann AC, Kossiakoff AA, Trent MS, Stansfeld PJ, Mancia F

EMDB-25834: 
CryoEM Structure of sFab COP-2 Complex with human claudin-4 and Clostridium perfringens enterotoxin C-terminal domain
Method: single particle / : Vecchio AJ, Orlando BJ

EMDB-25835: 
CryoEM Structure of sFab COP-3 Complex with human claudin-4 and Clostridium perfringens enterotoxin C-terminal domain
Method: single particle / : Vecchio AJ, Orlando BJ

EMDB-23708: 
ATP-bound AMP-activated protein kinase
Method: single particle / : Yan Y, Mukherjee S, Harikumar KG, Strutzenberg T, Zhou XE, Powell SK, Xu T, Sheldon R, Lamp J, Brunzelle JS, Radziwon K, Ellis A, Novick SJ, Vega IE, Jones R, Miller LJ, Xu HE, Griffin PR, Kossiakoff AA, Melcher K

EMDB-13424: 
Structure of Multidrug and Toxin Compound Extrusion (MATE) transporter NorM by NabFab-fiducial assisted cryo-EM
Method: single particle / : Bloch JS, Mukherjee S, Kowal J, Niederer M, Pardon E, Steyaert J, Kossiakoff AA, Locher KP

EMDB-13425: 
Map of complex of Vibrio cholerae MATE transporter NorM, chimeric nanobody NorM-Nb17_4, NabFab, and anti-Fab nanobody with fulcrum in center of micelle
Method: single particle / : Bloch JS, Mukherjee S, Kowal J, Niederer M, Pardon E, Steyaert J, Kossiakoff AA, Locher KP

EMDB-13426: 
Structure of homo-dimeric Staphylococcus capitis divalent metal ion transporter (DMT) by NabFab-fiducial assisted cryo-EM
Method: single particle / : Bloch JS, Mukherjee S, Kowal J, Kossiakoff AA, Locher KP
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
